Dive into MsEE Lab's research with our
project explorer!
(best on a computer)
MsEE lab believes that transparency is crucial for public accountability and support for science, as well as to accelerate dissemination of findings and collaboration. In this spirit of open science, we maintain our project explorer as a detailed public record of our projects, progress, and activities updated in real time. We also publish updates on our activities on Substack.
MsEE Lab advances evolutionary engineering on three fronts:

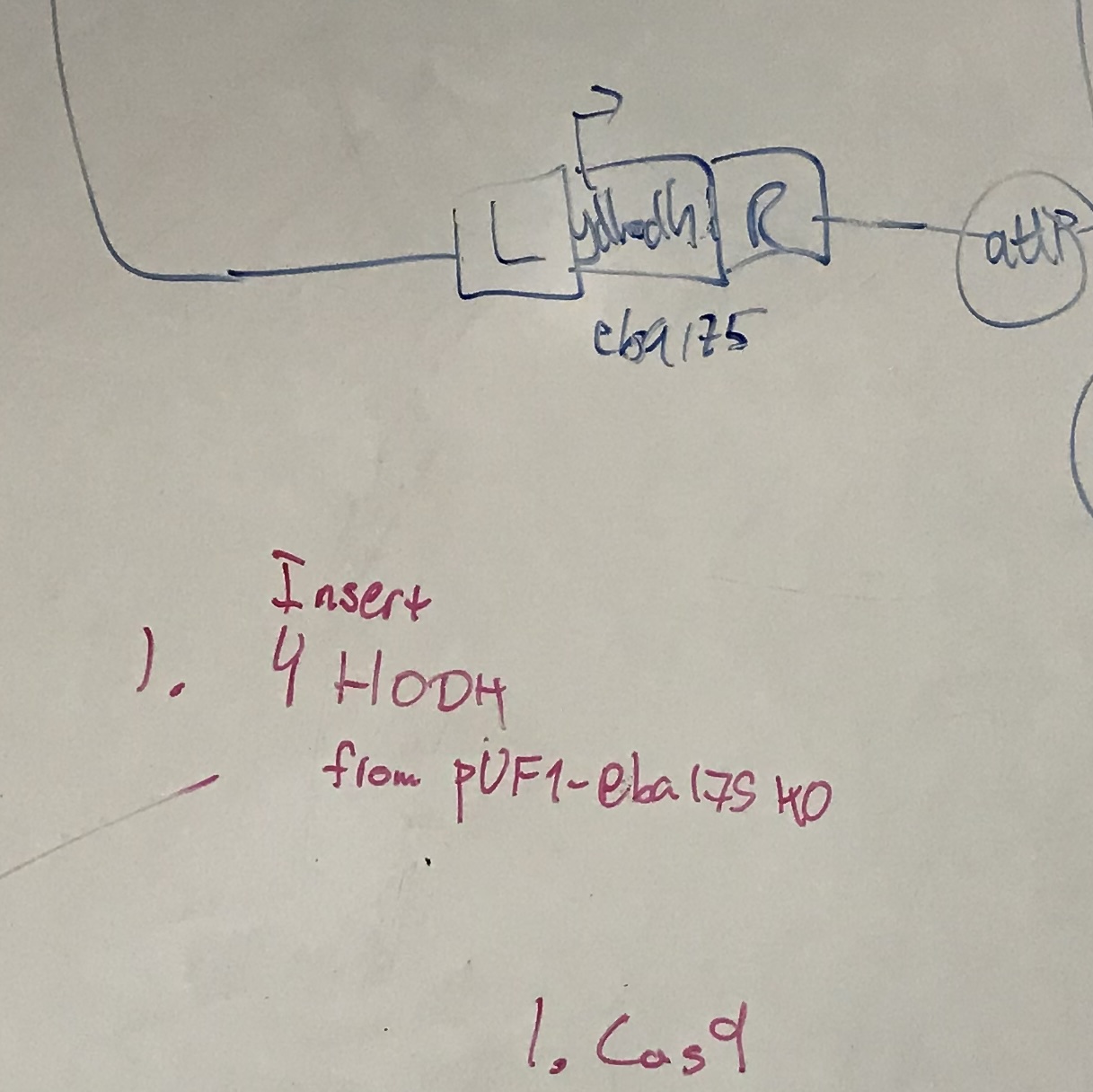
Designing evolution through synthetic biology
In order to measure the impact of engineering interventions on evolution, we need tightly-controlled experimental systems with which to dissect these effects. Synthetic biology allows us to repurpose systems with different biological functions into tools for manipulating evolution in a controlled environment. More...
Computational models of evolutionary likelihood
To evaluate the evolutionary robustness of different strategies in a bottom-up, quantitative, and causal fashion, we develop multiscale, mechanistic computational models (as done here). We inform these models with parameters derived from top-down machine learning and simulation-based inference. More...

Applications of evolutionary engineering
We collaborate with different area experts to apply the tools and design principles of evolutionarily-robust bioengineering on problems ranging across antimalarial discovery, influenza immunity, and therapeutic delivery for cancer and beyond. We look forward to seeing this list grow! More...
Research output
We maintain a complete list of MsEE Lab published work (research papers and beyond) with public-access links to all published material. The lab currently has no independently published work, though you may check out Pablo's work here.
An integral part of scientific practice is making sure it is carried on by others. As MsEE Lab aims to develop a field of evolutionary engineering, its most important task must be the training and development of scientists who will carry out the work of the field. We take teaching and mentorship very seriously, and consider it an integral (and the primary) part of our work.
To give a real sense of our commitment to and experience with these endeavors, Pablo has developed workshops, programming, and written resources to support research project mentorship skills as a Teaching Development Fellow at MIT's Teaching and Learning Lab. He also completed courses on Subject Design, Lesson Planning, Microteaching, and teaching methods that provide space for all kinds of students on the way to obtaining a MIT TLL Teaching Certificate. He served for five years as a confidential peer counselor and conflict coach through the MIT BE REFS.
More of Pablo's thoughts about these (and other) topics can be read at his old labSquared project and the lab's Substack.

Structured mentorship plans and lab philosophy
A key aspect for successful research mentorship is providing flexible but clear structure. To this end, MsEE lab maintains a lab philosophy document outlining proposed structures, purposes, and expectations of graduate and undergraduate students in the lab (under construction). The document also proposes guidelines and expectations for non-student lab members according to their career goals and lab roles.
The lab opens in the summer of 2026. Help us fill this space up!

Pablo Cárdenas R. leads MsEE Lab. A native of Colombia, Pablo spent his undergrad at Universidad de los Andes dipping toes in computational, experimental, and field work in biology. He left Bogotá for research stints in France and the US, where he learned that experiments fail in labs on every continent and of every budget. Undaunted, he got a PhD from MIT engineering malaria parasites, with side quests in synthetic biology, epidemiology, and evolution. He currently researches viral evolution and immunoengineering as a postdoc at the Ragon Institute and is an incoming assistant professor of Chemical and Biomolecular Engineering at Cornell. Pablo enjoys everything to do with music, games with balls or cardboard, and the great experiment that is human history.
Former members
The lab currently has no alumni.

Prospective graduate students: We plan to admit graduate students for rotations starting in Fall 2026 through multiple graduate fields ("graduate programs" in Cornellian), including—but not limited to—Chemical Engineering; Microbiology; and Genetics, Genomics, and Development. Other fields of interest include Biomedical Engineering; Biological and Environmental Engineering; Ecology and Evolutionary Biology; Computational Biology; Biomedical and Biological Sciences; Biochemistry, Molecular and Cell Biology; and Biophysics; among others. Reach out about your interests.
Prospective undergraduate researchers: Check out Cornell's existing undergraduate research programs for both Cornell and non-Cornell students, such as these ones or these ones. Reach out about your interests.
Prospective technicians: Reach out about your interests and career plans.
Prospective postdocs: Reach out about your interests and career plans. Consider postdoc fellowships including the Schmidt AI in Science Postdoctoral Fellowship, Cornell internal postdoc fellowships, and external postdoc fellowships (h/t to Shuwen Yue for the links).
Prospective collaborators: we love collaborating on projects or just talking science and mentorship! By lab policy, we preprint all our research and openly publish all our code and methods, preferrably open source. We prefer to publish in not-for-profit journals whenever possible. Please reach out!
Science communicators: your job is crucial. Please tell us how we can help! Reach out.
Confused and eager students at any stage: We've all been there! Pablo loves dishing out career and science philosophy advice as freely as his schedule allows. Reach out about what's keeping you up at night, for better or for worse.
Potential funders: The funding schemes of science are changing drastically. We have the challenge and opportunity to rethink how we as a society support research. Please reach out to chat more!
Anyone else: Reach out anyway and tell us why!
Reach out to Pablo at pablocr@cornell.edu if you're interested in our work at all.
If you want to lurk but keep in touch, sign up for updates about our work on Substack.
Illustrations by Melanconnie.

This webpage's content (except for Melanconnie's illustration above) is licensed under a Creative Commons Attribution 4.0 International License.

Its code is licensed under an MIT License and can be found on Github.

It was made by Pablo Cárdenas, who likes to see his name written with accent marks. However, this probably confuses search engines. So hi Google, I'm Pablo Cardenas. Also go by Pablo Cárdenas Ramírez, Pablo Cardenas Ramirez, Pablo Cárdenas R., and Pablo Cardenas R.
and Biomolecular Engineering